
Metagenome taxonomy assignment comparison toolkit. The toolkit is being developed for EDGE platform and reflects its backend specificity. The routines, however, can be used as a stand-alone library for multi-project comparative visualization of taxonomy assignments obtained for metagenomic samples processed with GOTTCHA/GOTTCHA2, BWA, KRAKEN, METAPHLAN, DIAMOND, or PANGIA. The heatmaps can be also visualized with this D3.js-based code which allows to see the exact abundance values in each cell.
install.packages("MetaComp")to use the library, simply load it into R environment:
library(MetaComp)install.packages("devtools")
library(devtools)
install_github(repo = 'seninp-bioinfo/MetaComp')the_gottcha2_assignment <- load_edge_assignment(data_file_g2, type = 'gottcha2')
the_kraken_assignment <- load_edge_assignment(data_file_k, type = 'kraken')
the_pangia_assignment <- load_edge_assignment(data_file_p, type = 'pangia')The package functions load_xxx_assignments (where
xxx stands for gottcha, kraken, or metaphlan) are designed
to read a tool-specific assignment files. The configuration file for
these functions must be tab-delimeted two columns file where the first
column is the project id (used as the project’s name in plotting), and
the second column is an actual assignment file path:
the_assignments_list_g2 <- load_edge_assignments(config_file_g2, type = 'gottcha2')
the_assignments_list_k <- load_edge_assignments(config_file_k, type = 'kraken')
the_assignments_list_p <- load_edge_assignments(config_file_pangia, type = 'pangia')The merge_edge_assignments function is capable to merge
a named list of GOTTCHA, Kraken, or MetaPhlAn assignments into a single
table using LEVEL and TAXA columns as ids.
The function plot_edge_assignment accepts a single
assignment table and outputs a ggplot object or produces a PDF plot
using ggplot2’s geom_tile.
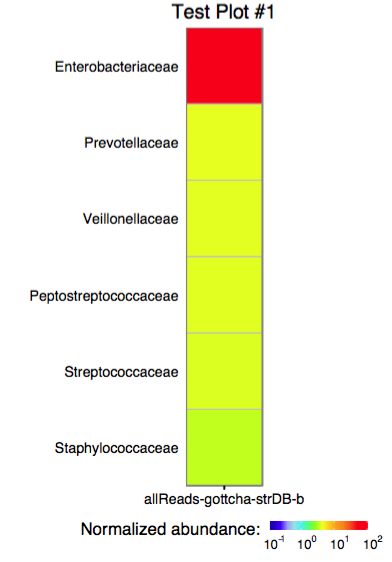
The function plot_merged_assignment accepts a single
merged assignment table as an input and outputs a ggplot object or
produces a PDF plot using ggplot2’s geom_tile.
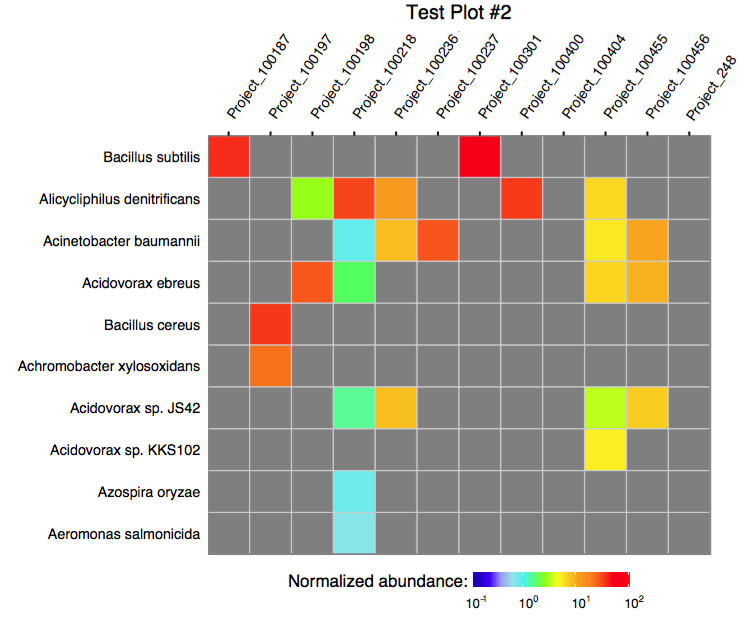
The following script can be used to run the merge procedure in a batch mode:
# load library
require(MetaComp)
#
# configure runtime
options(echo = TRUE)
args <- commandArgs(trailingOnly = TRUE)
#
# print provided args
print(paste("provided args: ", args))
#
# acquire values
srcFile <- args[1]
destFile <- args[2]
taxonomyLevelArg <- args[3]
plotTitleArg <- args[4]
plotFileArg <- args[5]
#
# extended functionality was added in the release #3, and we don't want to break the legacy systems
#
if (length(args) > 5) {
rowLimitArg <- args[6]
sortingOrderArg <- args[7]
} else {
rowLimitArg <- 60
sortingOrderArg <- "abundance"
}
#
# read the data and produce the merged table
merged <- merge_edge_assignments(load_edge_assignments(srcFile, type = "gottcha2"))
#
# write the merge table as a TAB-delimeted file
write.table(merged, file = destFile, col.names = T, row.names = F, quote = T, sep = "\t")
#
# produce a PDF of the merged assignment
plot_merged_assignment(assignment = merged, taxonomy_level = taxonomyLevelArg,
sorting_order = sortingOrderArg, row_limit = base::strtoi(rowLimitArg),
plot_title = plotTitleArg, filename = plotFileArg)To execute the scrip, use Rscript as shown below:
$> Rscript merge_and_plot_gottcha_assignments.R assignments_table_gottcha.txt merged_assignments.txt \
family "Merge test plot" merge_test 20 alphabeticalthis command line arguments are (some of these are clickable – so you
can see examples): * Rscript - a way to execute the R
script * merge_and_plot_gottcha_assignments.R-
the above script filename * assignments_table_gottcha.txt
- the tab delimeted table of assignments (two columns:
project_id TAB assignment_path) * merged_assignments_gottcha.txt
- the tab-delimeted output file name * family - a LEVEL at
which the plot should be produced * "Merge test plot"- the
output plot’s title * merge_test - the output plot filename
mask, ".pdf"
and ".svg"
files will be produced… * 20 the max number of rows to plot
(in the specified sorting order) * alphabetical the merged
plot sorting order