
The goal of ezcox is to operate a batch of univariate or multivariate Cox models and return tidy result.
You can install the released version of ezcox from CRAN with:
install.packages("ezcox")And the development version from GitHub with:
# install.packages("remotes")
remotes::install_github("ShixiangWang/ezcox")It is possible to install ezcox from Conda
conda-forge channel:
conda install r-ezcox --channel conda-forgeVisualization feature of ezcox needs the recent version of forestmodel, please run the following commands:
remotes::install_github("ShixiangWang/forestmodel")This is a basic example which shows you how to get result from a batch of cox models.
library(ezcox)
#> Welcome to 'ezcox' package!
#> =======================================================================
#> You are using ezcox version 1.0.2
#>
#> Project home : https://github.com/ShixiangWang/ezcox
#> Documentation: https://shixiangwang.github.io/ezcox
#> Cite as : arXiv:2110.14232
#> =======================================================================
#>
library(survival)
# Build unvariable models
ezcox(lung, covariates = c("age", "sex", "ph.ecog"))
#> => Processing variable age
#> ==> Building Surv object...
#> ==> Building Cox model...
#> ==> Done.
#> => Processing variable sex
#> ==> Building Surv object...
#> ==> Building Cox model...
#> ==> Done.
#> => Processing variable ph.ecog
#> ==> Building Surv object...
#> ==> Building Cox model...
#> ==> Done.
#> # A tibble: 3 × 12
#> Variable is_cont…¹ contr…² ref_l…³ n_con…⁴ n_ref beta HR lower…⁵ upper…⁶
#> <chr> <lgl> <chr> <chr> <int> <int> <dbl> <dbl> <dbl> <dbl>
#> 1 age FALSE age age 228 228 0.0187 1.02 1 1.04
#> 2 sex FALSE sex sex 228 228 -0.531 0.588 0.424 0.816
#> 3 ph.ecog FALSE ph.ecog ph.ecog 227 227 0.476 1.61 1.29 2.01
#> # … with 2 more variables: p.value <dbl>, global.pval <dbl>, and abbreviated
#> # variable names ¹is_control, ²contrast_level, ³ref_level, ⁴n_contrast,
#> # ⁵lower_95, ⁶upper_95
# Build multi-variable models
# Control variable 'age'
ezcox(lung, covariates = c("sex", "ph.ecog"), controls = "age")
#> => Processing variable sex
#> ==> Building Surv object...
#> ==> Building Cox model...
#> ==> Done.
#> => Processing variable ph.ecog
#> ==> Building Surv object...
#> ==> Building Cox model...
#> ==> Done.
#> # A tibble: 4 × 12
#> Variable is_cont…¹ contr…² ref_l…³ n_con…⁴ n_ref beta HR lower…⁵ upper…⁶
#> <chr> <lgl> <chr> <chr> <int> <int> <dbl> <dbl> <dbl> <dbl>
#> 1 sex FALSE sex sex 228 228 -0.513 0.599 0.431 0.831
#> 2 sex TRUE age age 228 228 0.017 1.02 0.999 1.04
#> 3 ph.ecog FALSE ph.ecog ph.ecog 227 227 0.443 1.56 1.24 1.96
#> 4 ph.ecog TRUE age age 228 228 0.0113 1.01 0.993 1.03
#> # … with 2 more variables: p.value <dbl>, global.pval <dbl>, and abbreviated
#> # variable names ¹is_control, ²contrast_level, ³ref_level, ⁴n_contrast,
#> # ⁵lower_95, ⁶upper_95lung$ph.ecog = factor(lung$ph.ecog)
zz = ezcox(lung, covariates = c("sex", "ph.ecog"), controls = "age", return_models=TRUE)
#> => Processing variable sex
#> ==> Building Surv object...
#> ==> Building Cox model...
#> ==> Done.
#> => Processing variable ph.ecog
#> ==> Building Surv object...
#> ==> Building Cox model...
#> ==> Done.
mds = get_models(zz)
str(mds, max.level = 1)
#> List of 2
#> $ Surv ~ sex + age :List of 19
#> ..- attr(*, "class")= chr "coxph"
#> ..- attr(*, "Variable")= chr "sex"
#> $ Surv ~ ph.ecog + age:List of 22
#> ..- attr(*, "class")= chr "coxph"
#> ..- attr(*, "Variable")= chr "ph.ecog"
#> - attr(*, "class")= chr [1:2] "ezcox_models" "list"
#> - attr(*, "has_control")= logi TRUE
show_models(mds)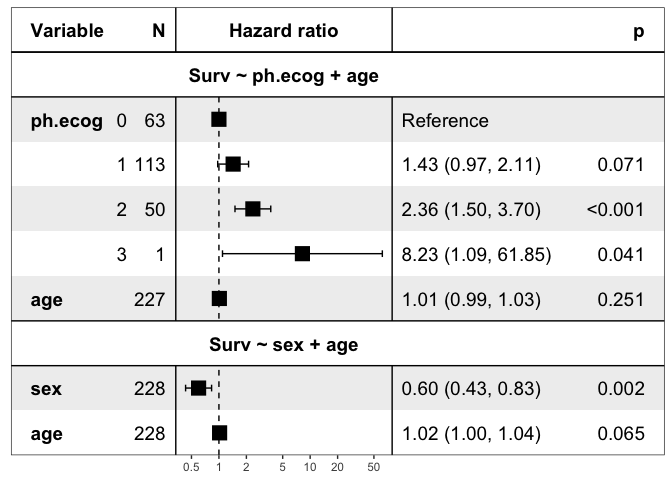
If you are using it in academic research, please cite the preprint arXiv:2110.14232 along with URL of this repo.