

Within light there is darkness, but do not try to understand that darkness. Within darkness there is light, but do not look for that light. Light and darkness are a pair like the foot before and the foot behind in walking.
– From the Zen teaching poem Sandokai.
The goal of halfmoon is to cultivate balance in propensity score models.
You can install the most recent version of halfmoon from CRAN with:
install.packages("halfmoon")You can also install the development version of halfmoon from GitHub with:
# install.packages("devtools")
devtools::install_github("malcolmbarrett/halfmoon")halfmoon includes several techniques for assessing the balance created by propensity score weights.
library(halfmoon)
library(ggplot2)
# weighted mirrored histograms
ggplot(nhefs_weights, aes(.fitted)) +
geom_mirror_histogram(
aes(group = qsmk),
bins = 50
) +
geom_mirror_histogram(
aes(fill = qsmk, weight = w_ate),
bins = 50,
alpha = 0.5
) + scale_y_continuous(labels = abs)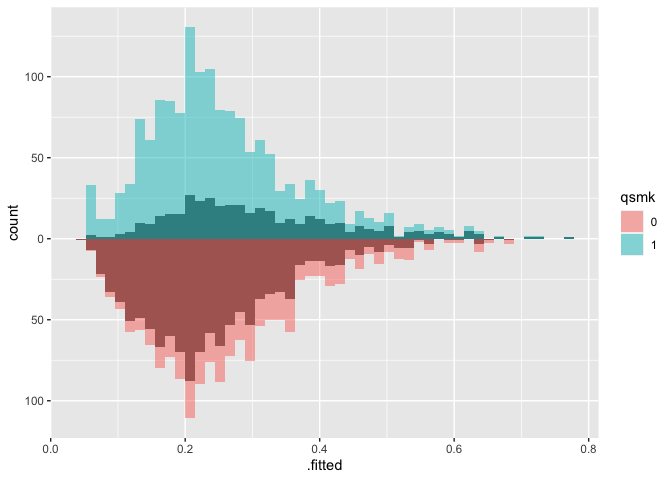
# weighted ecdf
ggplot(
nhefs_weights,
aes(x = smokeyrs, color = qsmk)
) +
geom_ecdf(aes(weights = w_ato)) +
xlab("Smoking Years") +
ylab("Proportion <= x")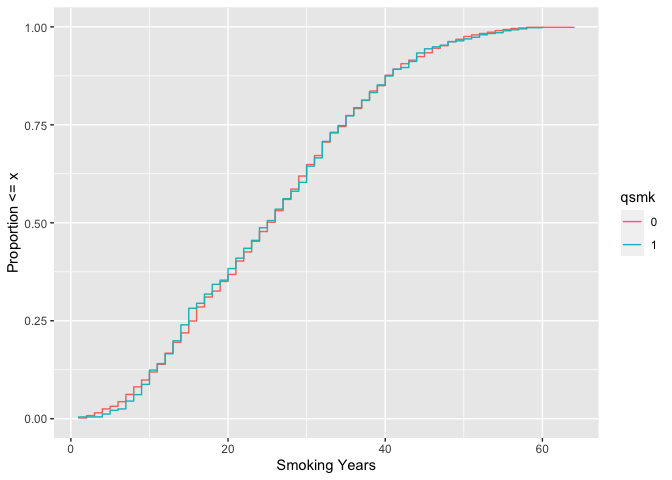
# weighted SMDs
plot_df <- tidy_smd(
nhefs_weights,
race:active,
.group = qsmk,
.wts = starts_with("w_")
)
ggplot(
plot_df,
aes(
x = abs(smd),
y = variable,
group = method,
color = method
)
) +
geom_love()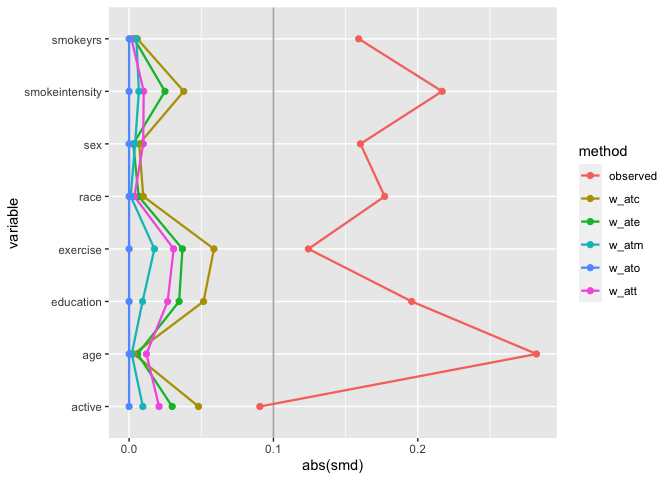
halfmoon also has support for working with matched datasets. Consider these two objects from the MatchIt documentation:
library(MatchIt)
# Default: 1:1 NN PS matching w/o replacement
m.out1 <- matchit(treat ~ age + educ + race + nodegree +
married + re74 + re75, data = lalonde)
# 1:1 NN Mahalanobis distance matching w/ replacement and
# exact matching on married and race
m.out2 <- matchit(treat ~ age + educ + race + nodegree +
married + re74 + re75, data = lalonde,
distance = "mahalanobis", replace = TRUE,
exact = ~ married + race)One option is to just look at the matched dataset with halfmoon:
matched_data <- get_matches(m.out1)
match_smd <- tidy_smd(
matched_data,
c(age, educ, race, nodegree, married, re74, re75),
.group = treat
)
love_plot(match_smd)
The downside here is that you can’t compare multiple matching
strategies to the observed dataset; the label on the plot is also wrong.
halfmoon comes with a helper function, bind_matches(), that
creates a dataset more appropriate for this task:
matches <- bind_matches(lalonde, m.out1, m.out2)
head(matches)
#> treat age educ race married nodegree re74 re75 re78 m.out1 m.out2
#> NSW1 1 37 11 black 1 1 0 0 9930.0460 1 1
#> NSW2 1 22 9 hispan 0 1 0 0 3595.8940 1 1
#> NSW3 1 30 12 black 0 0 0 0 24909.4500 1 1
#> NSW4 1 27 11 black 0 1 0 0 7506.1460 1 1
#> NSW5 1 33 8 black 0 1 0 0 289.7899 1 1
#> NSW6 1 22 9 black 0 1 0 0 4056.4940 1 1matches includes an binary variable for each
matchit object which indicates if the row was included in
the match or not. Since downweighting to 0 is equivalent to filtering
the datasets to the matches, we can more easily compare multiple matched
datasets with .wts:
many_matched_smds <- tidy_smd(
matches,
c(age, educ, race, nodegree, married, re74, re75),
.group = treat,
.wts = c(m.out1, m.out2)
)
love_plot(many_matched_smds)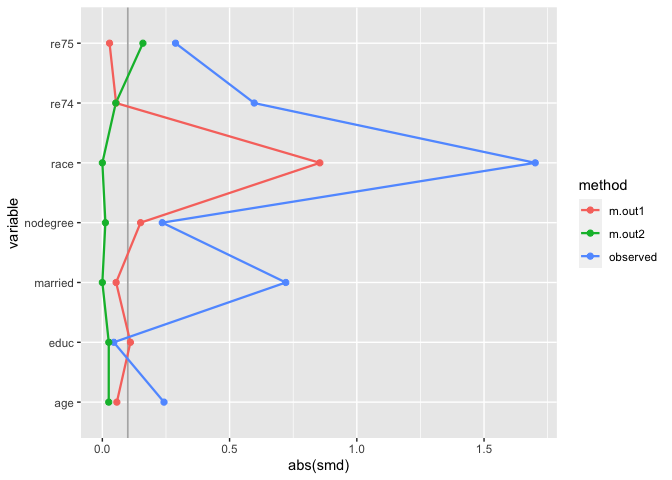
We can also extend the idea that matching indicators are weights to weighted mirrored histograms, giving us a good idea of the range of propensity scores that are being removed from the dataset.
# use the distance as the propensity score
matches$ps <- m.out1$distance
ggplot(matches, aes(ps)) +
geom_mirror_histogram(
aes(group = factor(treat)),
bins = 50
) +
geom_mirror_histogram(
aes(fill = factor(treat), weight = m.out1),
bins = 50,
alpha = 0.5
) + scale_y_continuous(labels = abs)