The goal of kerntools is to provide R tools for working
with a family of Machine Learning methods called kernel methods. It can
be used to complement other R packages like kernlab. Right
now, kerntools implements several kernel functions for
treating non-negative and real vectors, real matrices, categorical and
ordinal variables, sets, and strings. Several tools for studying the
resulting kernel matrix or to compare two kernel matrices are available.
These diagnostic tools may be used to infer the kernel(s) matrix(ces)
suitability in training models. This package also provides functions for
computing the feature importance of Support Vector Machines (SVMs)
models, and display customizable kernel Principal Components Analysis
(PCA) plots. For convenience, widespread performance measures and
feature importance barplots are available for the user.
If you want to see a real-life application of kerntools,
you can check the following paper:
Installing kerntools is easy. In the R console:
install.packages("kerntools")Once the package is installed, it can be loaded anytime typing:
library(kerntools)kerntools requires R (>= 2.10). Currently, it also
relies on the following packages:
dplyrggplot2kernlabmethodsreshape2stringiUsually, if some of these packages are missing in your library, they
will be installed automatically when kerntools is
installed.
Imagine that you want to perform a (kernel) PCA plot but your dataset
consist of categorical variables. This can be done very easily with
kerntools! First, you chose an appropriate kernel for your
data (in this example, the Dirac kernel for categorical variables), and
then you pass the output of the Dirac() function to the
kPCA() function.
head(showdata)
#> Favorite.color Favorite.actress Favorite.actor Favorite.show
#> 1 red Sophie Turner Josh O'Connor The crown
#> 2 black Soo Ye-jin Hyun Bin Bridgerton
#> 3 red Lorraine Ashbourne Henry Cavill Bridgerton
#> 4 blue Sophie Turner Alvaro Morte La casa de papel
#> 5 red Sophie Turner Michael K Williams The wire
#> 6 yellow Sophie Turner Kit Harington Game of Thrones
#> Liked.new.show
#> 1 Yes
#> 2 No
#> 3 Yes
#> 4 No
#> 5 Yes
#> 6 No
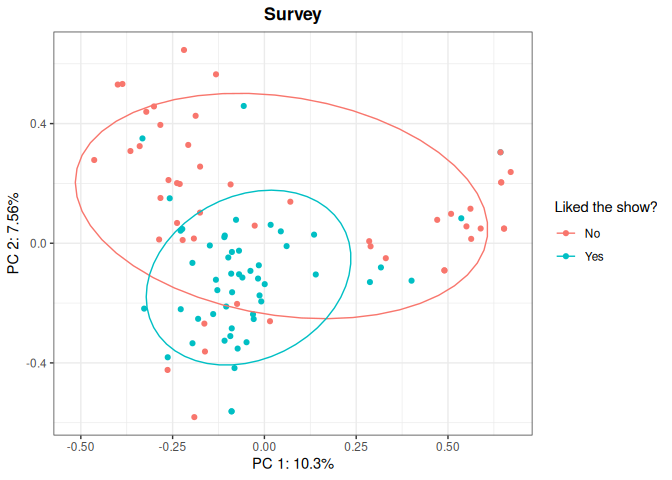
KD <- Dirac(showdata[,1:4])
dirac_kpca <- kPCA(KD,plot=c(1,2),title="Survey", name_leg = "Liked the show?",
y=showdata$Liked.new.show, ellipse=0.66)
dirac_kpca$plot
You can customize your kernel PCA plot: apart from picking which
principal components you want to display (in the example: PC1 and PC2),
you may want to add a title, or a legend, or use different colors to
represent an additional variable of interest, so you can check patterns
on your data. To see in detail how to customize a kPCA()
plot, please refer to the documentation. The projection matrix is also
returned (dirac_kpca$projection), so you may use it for
further analyses and/or creating your own plot.
Right now, kerntools can deal effortlessly with the
following kinds of data:
Several tools for visualizing and comparing kernel matrices are provided.
Regarding kernel PCA, kerntools allows the user to:
kerntools or provided by the user.When using some specific kernels, kerntools computes the
importance of each variable or feature in a Support Vector Machine (SVM)
model. kerntools does not train SVMs or other prediction
models, but it can recover the feature importance of models fitted with
other packages (for instance kernlab). These importances
can be sorted and summarized in a customizable barplot.
Finally, the following performance measures for regression, binary and multi-class classification are implemented:
kerntools contains a categorical toy dataset called
showdata and a real-world count dataset called
soil.
To see detailed and step-by-step examples that illustrate the main
cases of use of kerntools, please have a look to the
vignettes:
browseVignettes(kerntools)The basic vignette covers the typical kerntools
workflow. Thorough documentation about the kernel functions implemented
in this package is in the “Kernel functions” vignette. If you want
instead to know more about kernel PCA and Coinertia analysis, you can
refer to the corresponding vignette too.
Remember that detailed, argument-by-argument documentation is available for each function:
help(kPCA) ## or the specific name of the function
?kPCAThe documentation of the example datasets is available in an analogous way, typing:
help(showdata)
?showdataTo know more about kernel functions, matrices and methods, you can consult the following reference materials:
Bishop, C. M., & Nasrabadi, N. M. (2006). Pattern recognition and machine learning (Vol. 4, No. 4, p. 738). Chapter 6, pp. 291-323. New York: springer.
Müller, K. R., Mika, S., Tsuda, K., & Schölkopf, K. (2018) An introduction to kernel-based learning algorithms. In Handbook of neural network signal processing (pp. 4-1). CRC Press.
Shawe-Taylor, J., & Cristianini, N. (2004). Kernel methods for pattern analysis. Cambridge university press.