

rbmiUtils bridges rbmi analysis results into
publication-ready regulatory tables and forest plots. It extends rbmi
for clinical trial workflows, providing additional utilities from data
validation, storing imputed data sets, through to formatted efficacy
outputs.
You can install the package from CRAN or the development version from GitHub:
| Type | Source | Command |
|---|---|---|
| Release | CRAN | install.packages("rbmiUtils") |
| Development | GitHub | remotes::install_github("openpharma/rbmiUtils") |
rbmiUtils provides additional support for the rbmi pipeline from raw
data to publication-ready outputs. Here is an example workflow to
illustrate these utilities using the bundled ADEFF
dataset:
library(rbmiUtils)
library(rbmi)
library(dplyr)
# Load example efficacy dataset and prepare factors
data("ADEFF", package = "rbmiUtils")
ADEFF <- ADEFF |>
mutate(
TRT = factor(TRT01P, levels = c("Placebo", "Drug A")),
USUBJID = factor(USUBJID),
AVISIT = factor(AVISIT, levels = c("Week 24", "Week 48"))
)
# Define analysis variables
vars <- set_vars(
subjid = "USUBJID",
visit = "AVISIT",
group = "TRT",
outcome = "CHG",
covariates = c("BASE", "STRATA", "REGION")
)
# Configure Bayesian imputation method
method <- method_bayes(
n_samples = 100,
control = control_bayes(warmup = 200, thin = 2)
)
# Step 1: Fit imputation model (draws)
dat <- ADEFF |> select(USUBJID, STRATA, REGION, TRT, BASE, CHG, AVISIT)
draws_obj <- draws(data = dat, vars = vars, method = method)
# Step 2: Generate imputed datasets
impute_obj <- impute(
draws_obj,
references = c("Placebo" = "Placebo", "Drug A" = "Placebo")
)
# Step 3: Extract stacked imputed data
ADMI <- get_imputed_data(impute_obj)
# Modification of the complete data is possible (i.e. collapsing variables).
# Step 4: Analyse each imputed dataset
ana_obj <- analyse_mi_data(data = ADMI, vars = vars, method = method, fun = ancova)
# Step 5: Pool results using Rubin's rules
pool_obj <- pool(ana_obj)
# Publication-ready table
efficacy_table(pool_obj, arm_labels = c(ref = "Placebo", alt = "Drug A"))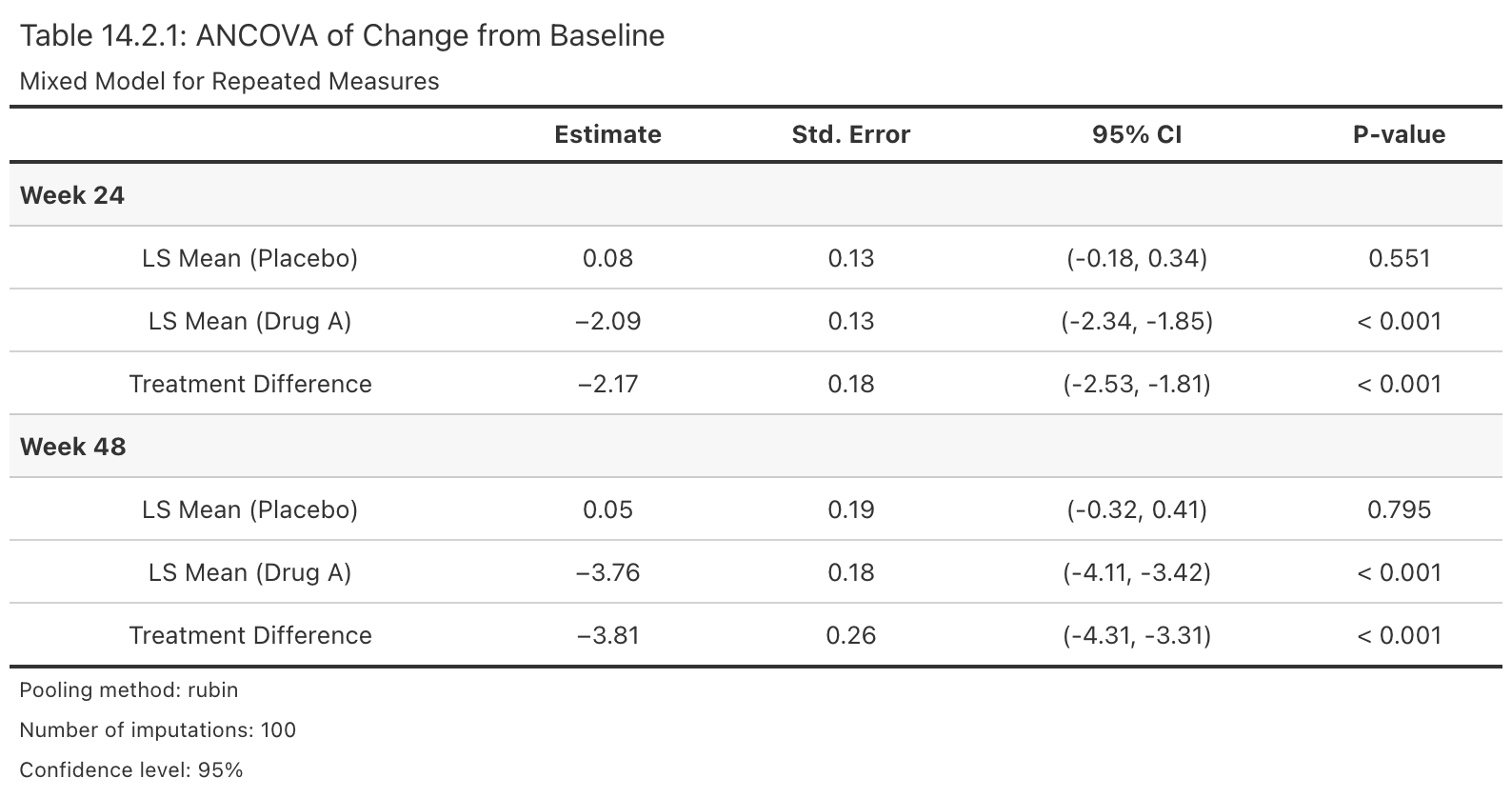
See the end-to-end pipeline vignette for the complete walkthrough from raw data to these outputs.
validate_data() – pre-flight checks on data structure
before imputationanalyse_mi_data() – run ANCOVA (or custom analysis)
across all imputationstidy_pool_obj() – tidy pooled results with visit-level
annotationsefficacy_table() – regulatory-style gt tables (ICH
Table 14.2.x format)plot_forest() – three-panel forest plots with
estimates, CIs, and p-valuespool_to_ard() – convert pool objects to pharmaverse ARD
format with optional MI diagnostic enrichment (FMI, lambda, RIV)get_imputed_data() – extract long-format imputed
datasetsdescribe_draws() – inspect draws objects (method,
samples, convergence diagnostics)describe_imputation() – inspect imputation objects
(method, M, missingness breakdown)format_pvalue() / format_estimate() –
publication-ready formattingThis package is experimental and under active development. Feedback and contributions are welcome via GitHub issues or pull requests.