

Simulate subject-level event reporting of clinical trial sites with the goal of detecting over- and under-reporting.
Monitoring reporting rates of subject-level clinical events (e.g. adverse events, protocol deviations) reported by clinical trial sites is an important aspect of risk-based quality monitoring strategy. Sites that are under-reporting or over-reporting events can be detected using bootstrap simulations during which patients are redistributed between sites. Site-specific distributions of event reporting rates are generated that are used to assign probabilities to the observed reporting rates.
The method is inspired by the ‘infer’ R package and Allen Downey’s blog article: “There is only one test!”.
install.packages("simaerep")You can install the development version from GitHub with:
# install.packages("devtools")
devtools::install_github("openpharma/simaerep")simaerep has been published as workproduct of the
Inter-Company Quality Analytics (IMPALA) consortium. IMPALA
aims to engage with Health Authorities inspectors on defining guiding
principles for the use of advanced analytics to complement, enhance and
accelerate current QA practices. simaerep has initially
been developed at Roche but is currently evaluated by other companies
across the industry to complement their quality assurance activities (see
testimonials).
Koneswarakantha, B., Adyanthaya, R., Emerson, J. et al. An Open-Source R Package for Detection of Adverse Events Under-Reporting in Clinical Trials: Implementation and Validation by the IMPALA (Inter coMPany quALity Analytics) Consortium. Ther Innov Regul Sci 58, 591–599 (2024). https://doi.org/10.1007/s43441-024-00631-8
Koneswarakantha, B., Barmaz, Y., Ménard, T. et al. Follow-up on the Use of Advanced Analytics for Clinical Quality Assurance: Bootstrap Resampling to Enhance Detection of Adverse Event Under-Reporting. Drug Saf (2020). https://doi.org/10.1007/s40264-020-01011-5
Download as pdf in the release section generated using thevalidatoR.
We have created an extension gsm.simaerep
so that simaerep event reporting probabilities can be added
to good statistical monitoring gsm.core
reports.
Calculate patient-level event reporting probabilities and the difference to the expected number of events on a simulated data set with 2 under-reporting sites.
suppressPackageStartupMessages(library(simaerep))
suppressPackageStartupMessages(library(dplyr))
suppressPackageStartupMessages(library(knitr))
set.seed(1)
df_visit <- sim_test_data_study(
n_pat = 1000, # number of patients in study
n_sites = 100, # number of sites in study
ratio_out = 0.02, # ratio of sites with outlier
factor_event_rate = -0.5, # rate of under-reporting
# non-constant event rates based on gamma distribution
event_rates = (dgamma(seq(1, 20, 0.5), shape = 5, rate = 2) * 5) + 0.1,
max_visit = 20,
max_visit_sd = 10,
study_id = "A"
)
df_visit %>%
select(study_id, site_id, patient_id, visit, n_event) %>%
head(25) %>%
knitr::kable()| study_id | site_id | patient_id | visit | n_event |
|---|---|---|---|---|
| A | S0001 | P000001 | 1 | 0 |
| A | S0001 | P000001 | 2 | 2 |
| A | S0001 | P000001 | 3 | 2 |
| A | S0001 | P000001 | 4 | 4 |
| A | S0001 | P000001 | 5 | 6 |
| A | S0001 | P000001 | 6 | 7 |
| A | S0001 | P000001 | 7 | 7 |
| A | S0001 | P000001 | 8 | 7 |
| A | S0001 | P000001 | 9 | 7 |
| A | S0001 | P000001 | 10 | 7 |
| A | S0001 | P000001 | 11 | 7 |
| A | S0001 | P000001 | 12 | 7 |
| A | S0001 | P000001 | 13 | 7 |
| A | S0001 | P000002 | 1 | 3 |
| A | S0001 | P000002 | 2 | 3 |
| A | S0001 | P000002 | 3 | 5 |
| A | S0001 | P000002 | 4 | 8 |
| A | S0001 | P000002 | 5 | 8 |
| A | S0001 | P000002 | 6 | 9 |
| A | S0001 | P000002 | 7 | 9 |
| A | S0001 | P000002 | 8 | 9 |
| A | S0001 | P000002 | 9 | 9 |
| A | S0001 | P000002 | 10 | 9 |
| A | S0001 | P000002 | 11 | 9 |
| A | S0001 | P000002 | 12 | 9 |
evrep <- simaerep(df_visit, mult_corr = TRUE)
plot(evrep, study = "A")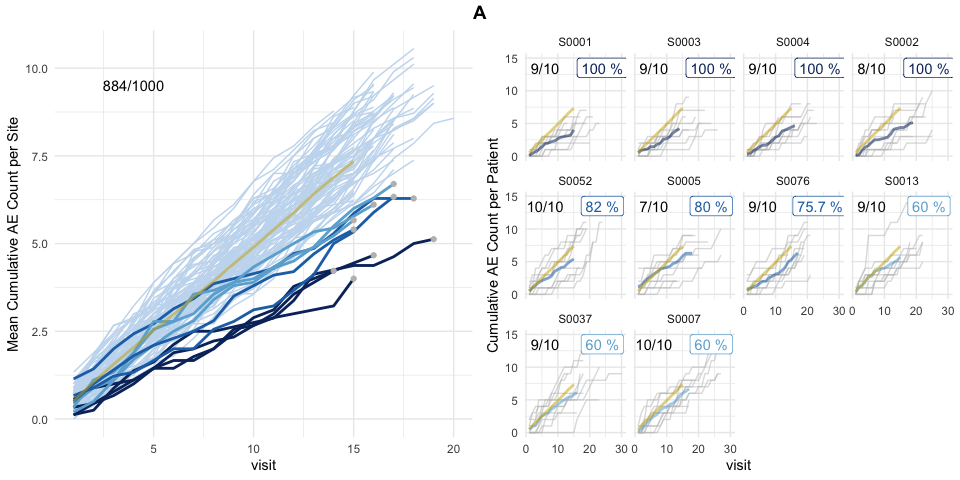
Left panel shows mean cumulative event reporting per site (blue lines) against mean cumulative event reporting of the entire study (golden line). Sites with either high under-reporting (negative probabilities) or high over-reporting (positive probabilities) are marked by grey dots and plotted in additional panels on the right. N denotes the number of sites. Right panel shows individual sites with total patient cumulative counts as grey lines. N denotes the number of patients, the percentage the under- and over-reporting probability and delta denotes the difference compared to the expected number of events.
The inframe
algorithm uses only dbplyr compatible table operations and
can be executed within a database backend as we demonstrate here using
duckdb.
However, we need to provide a in database table that has as many rows
as the desired replications in our simulation, instead of providing an
integer for the r parameter.
con <- DBI::dbConnect(duckdb::duckdb(), dbdir = ":memory:")
df_r <- tibble(rep = seq(1, 1000))
dplyr::copy_to(con, df_visit, "visit")
dplyr::copy_to(con, df_r, "r")
tbl_visit <- tbl(con, "visit")
tbl_r <- tbl(con, "r")
evrep <- simaerep(
tbl_visit,
r = tbl_r
)
plot(evrep, study = "A")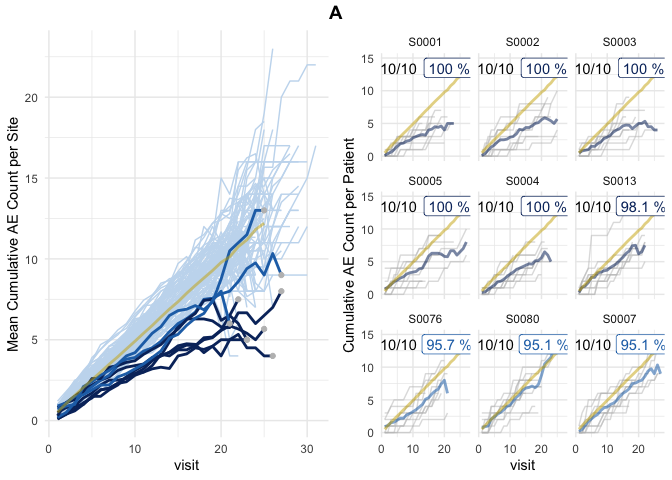
DBI::dbDisconnect(con)